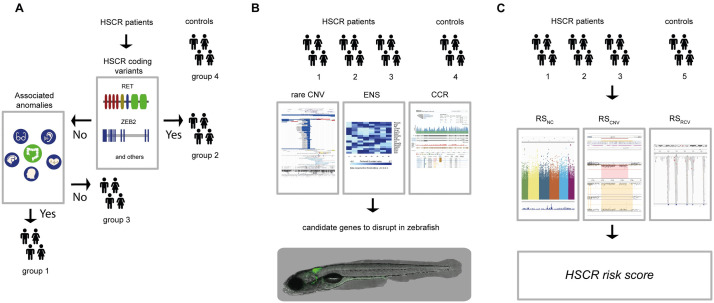
Fig 1
(A) For this study we included 326 controls (group 4) and 58 HSCR patients. We determined the RET and / or known disease gene coding mutations of HSCR patients (n = 58). The HSCR patients with coding mutations were included in group 2 (n = 15). We determined the presence of associated anomalies in the rest of the HSCR patients, including them in either the group containing associated anomalies (HSCR-AAM; group 1, n = 23) or in the group without associated anomalies (group 3, n = 30). (B) For all subgroups of HSCR patients and the controls Copy Number profiles were determined. To select candidate genes, we ranked CNVs according to their frequency in the unaffected controls from the Deciphering Developmental Disorders project (for illustrative purposes a screenshot of their UCSC genome browser track;