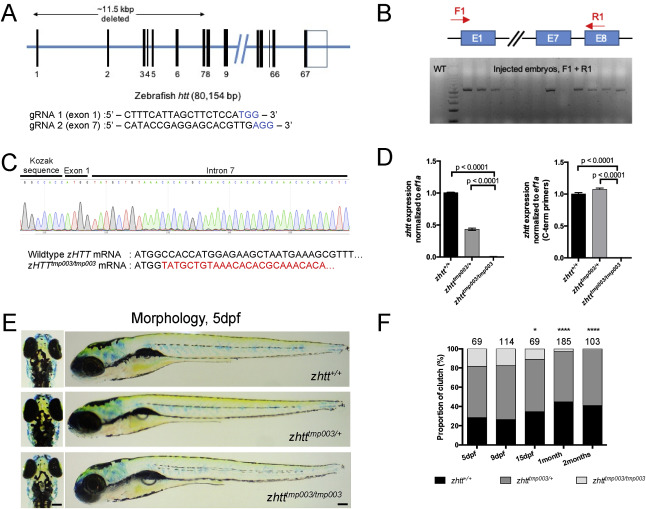
Fig. 3 Generation and characterization of HTT hemizygous and knockout zebrafish. (A) Graphical representation of zHTT genomic locus and sequences of sgRNAs used for CRISPR/Cas9 editing. Red box indicates the sequences that are deleted upon CRISPR/Cas9 cutting. (B) Distal primers F1/R1 are able to detect the presence of large genomic deletion on injected embryos. (C) Representative Sanger sequencing trace of injected embryo showing that large deletion removes a large portion of Exon 1 to Intron 7 of zHTT. Truncation removes a large chunk of sequence in zHTT mRNA beyond the first ATG codon. (D) qRT-PCR analysis of zhtt mRNA expression levels of 5dpf wildtype, hemizygous, and knockout zHTT embryos. Mean values ± SEM. n = 2 pools of 12 zebrafish for each genotype. p-values determined by one way ANOVA with Dunnett’s post hoc test. (E) Morphology of 5dpf zhtt wildtype, hemizygous, and knockout zebrafish. Lateral and dorsal views. Scale bar = 100μm. (F) Genotypic ratios of progenies from heterozygous intercrosses of zhtttmp003/+. Dotted lines represent expected ratio of each genotype. Number of fish analyzed per clutch is represented above the bars. zhtttmp003/tmp003 are significantly below expected ratio at 15dpf, 1 month, and 2 months. p-values determined by Chi-square analysis.*p = 0.0265; ****p<0.0001. dpf = days postfertilization.
Reprinted from Developmental Biology, 458(1), Sidik, H., Ang, C.J., Pouladi, M.A., Huntingtin confers fitness but is not embryonically essential in zebrafish development, 98-105, Copyright (2019) with permission from Elsevier. Full text @ Dev. Biol.