Fig. 2
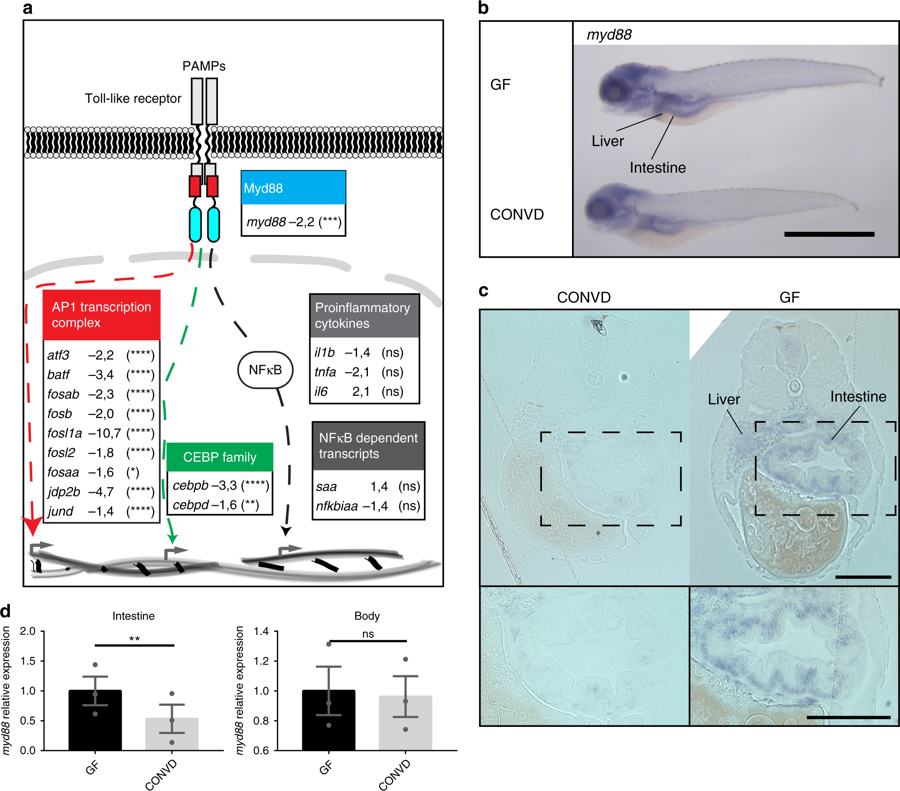
Microbes transcriptionally suppress Myd88 and downstream signaling components. a Graphic representation of the canonical TLR stimulated Myd88 dependent transcriptional signaling though the AP1 transcription complex (red), the CCAAT/enhancer binding protein family (green) or the NF-?B nuclear translocation (gray), along with associated gene names and fold-change values in the conventionalized group relative to germ-free. Nine transcription factors of the AP1 transcription complex and two members of the CCAAT/enhancer binding protein (C/EBP) family exhibited significant transcriptional suppression upon colonization. The activation of NF-?B is not readily observable by transcriptomics, however neither the NF-?B dependent transcripts serum amyloid A (saa) or NF-?B inhibitor Alpha a (nfkbiaa) or the proinflammatory cytokines normally associated with NF-?B activation were significantly regulated. It should be noted that the transcriptional foldchanges are derived from whole-body transcriptomics and that it cannot be concluded that the transcriptional changes represented here all take place in the same cells, even though they are part of the same regulatory pathway. Statistical evaluations represent the Benjamini-Hochberg adjusted P-values of the RNAseq data comparing conventionalized embryos to germ-free controls. *P???0.05; **P???0.01; ***P???0.001; ****P???0.0001; ns?=?not significant. b Whole-mount in situ hybridization reveals that myd88 is expressed primarily in the intestine and liver in 5 DPF embryos. Scalebar represents 1?mm. c 2?Ám transverse sections of plastic embedded in situ hybridization of myd88 comparing the pattern in a germ-free and a conventionalized embryo. Scalebar represents 100?Ám. d qPCR analysis comparing the relative expression levels of myd88 in germ-free versus conventionalized 5 DPF embryos in intestines versus body tissues, (mean?▒?s.e.m., n?=?3 biological replicates, 30 embryos per group), **P???0,01; by Student?s t-test