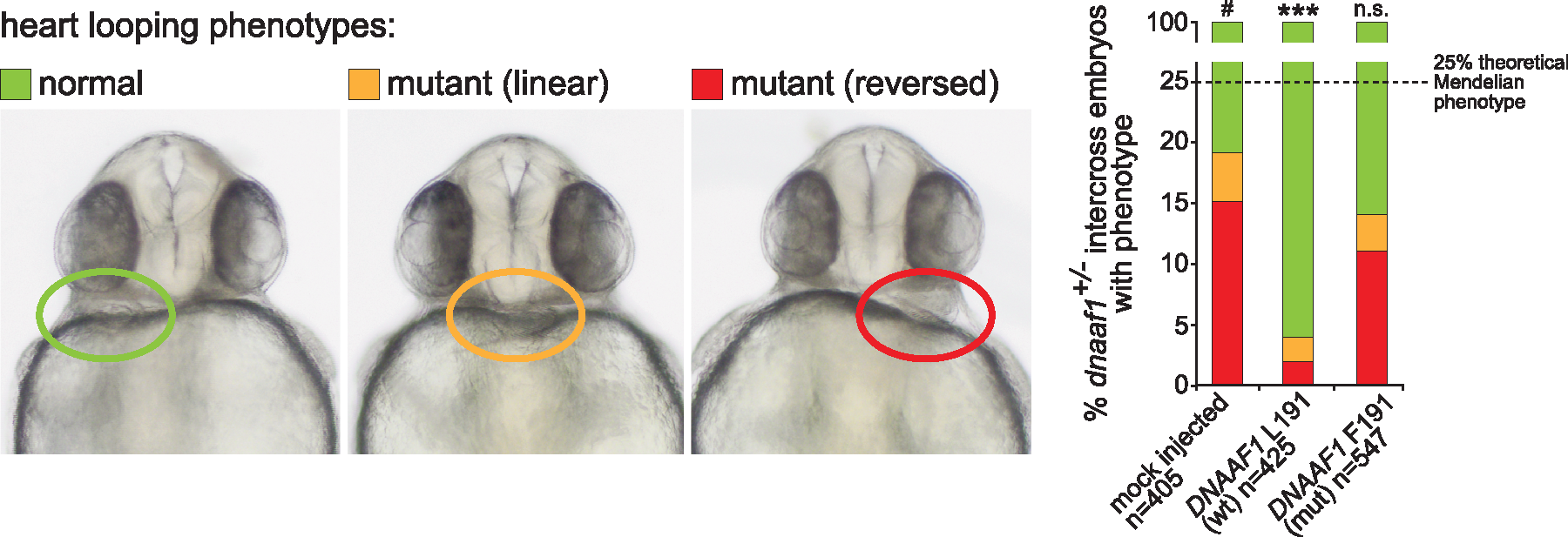
Fig. 2
Genetic complementation of heart looping defects in dnaaf1?/? mutant zebrafish embryos: Zebrafish embryos from dnaaf1+/? heterozygote-heterozygote inter-crosses were either mock injected (control, n?=?405 embryos), or injected with mRNA expressed from human wild-type (wt, n?=?425 embryos) and p.Leu191Phe-mutant (mut, n?=?547 embryos) pCS2+?DNAAF1 constructs. In all cases, a mixture of normal looping (green), reversed looping (red) or ?linear? looping indicating a lack of heart looping (orange) were seen and representative examples are shown at 72 h post fertilization (left panels). On the right, the bar graph quantifies the three positions of heart looping in all three experiments, showing that phenotypic rescue is seen after injection of wild-type but not mutant RNA. n.s.=?not significant; *** P?<?0.001 (one-way ANOVA, n?=?3 biological replicates). The dashed line indicates the 25% expected Mendelian ratio of embryos that should be homozygous mutant and manifest a phenotype.