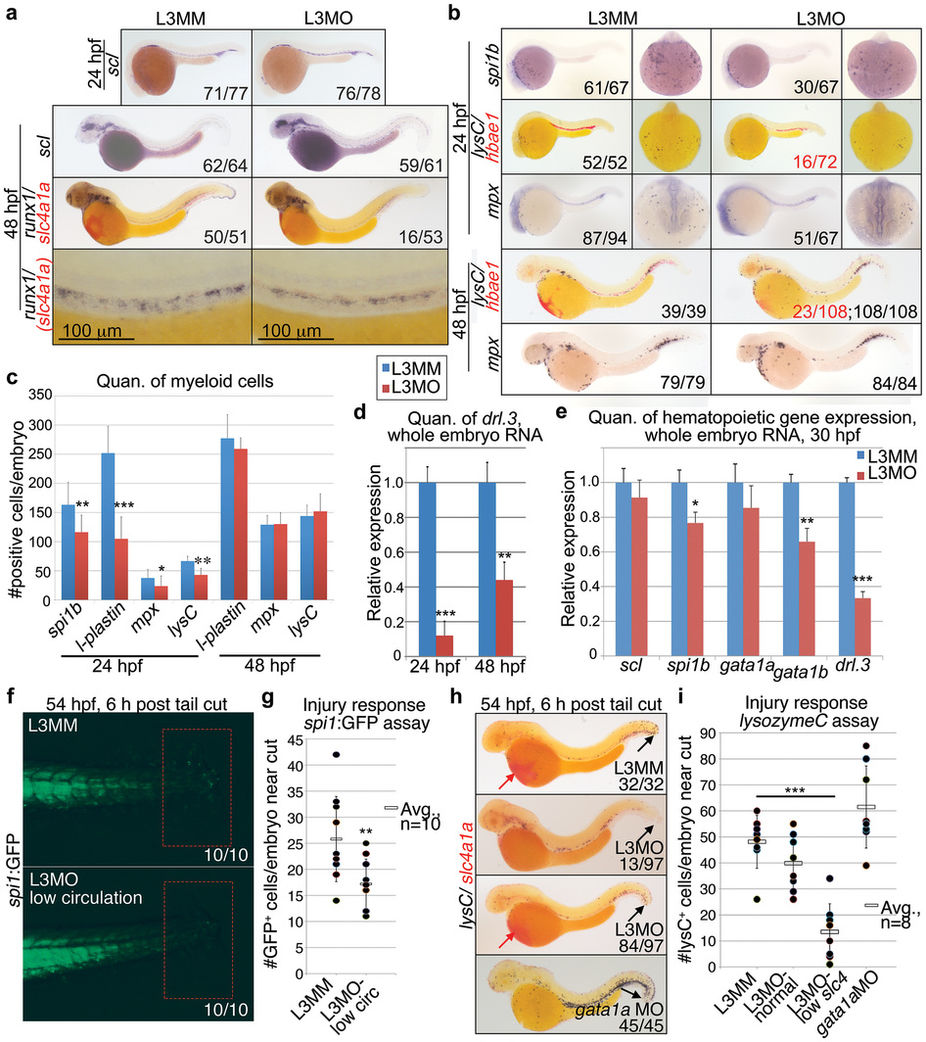
Fig. 4
Fig. 4
Knockdown of drl.3 transiently decreases myeloid cells without altering the emergence of primitive progenitor and definitive stem cells.
(a) WISH of scl at 24 and 48?hpf, and runx1 (dark blue)/slc4a1a (red) at 48?hpf in L3MM- and L3MO-injected embryos, as labeled. Embryos shown as lateral views. (b) WISH of spi1b, l-plastin and mpx at 24?hpf and l-plastin and mpx 48?hpf. Dorsal, anterior (24?hpf only, right panels) and lateral views are shown. (c) Quantitation of the number of the WISH spi1b+, l-plastin+ and mpx+ cells in the anterior of the embryo at 24?hpf and total body l-plastin+ and mpx+ cells at 48?hpf in L3MM (blue) and L3MO-injected (red) embryos (N = 8 for each column except for mpx at 24?hpf where N = 15, bars show mean ± S.E.). **P = 0.0044, ***P < 0.0001 and *P = 0.0156 (Student's t-test). (d) Quantitative real-time PCR analysis of drl.3 in whole embryo RNA samples from 24 and 48?hpf drl.3 morphants (red) and controls (L3MM, blue, set to 1, arbitrary units). **P = 0.0042 and ***P < 0.0001 (Student's t-test). (e) Quantitative real time PCR analysis of scl, spi1b, gata1a, gata1b, and drl.3 in whole embryo RNA samples from pools of 30?hpf drl.3 morphants (red) compared to control-injected embryos (blue, set to 1, arbitrary units). *P = 0.0114, **P = 0.0083 and ***P = 0.0005 (Student's t-test). (d?e) Bars show mean ± S.D., from three independent experiments. Expression was normalized to gapdh. (f) Tail region of spi1:GFP embryos at 54?hpf, 6?hours after tail transection. Selected L3MO embryos had decreased circulating cells; controls were randomly selected, and had normal circulation. Red boxes = tail cut region. (g) Number of spi1:GFP+ cells in tail cut region in control or drl.3 morphants. P = 0.007 (Student's t-test). (h) WISH of lysozymeC/lysC (blue) and slc4a1a (red) at 54?hpf, 6?hours after tails were cut. (i) Number of lysC+ cells in an equal sized region surrounding the tail in the indicated embryos at 54?hpf, 6?hours after tail cuts were performed. P = 6.77E-5, Student's t-test.