Fig. 1
Zebrafish cyclin Dx is a member of the cyclin D family, and zccndx is expressed during early development.
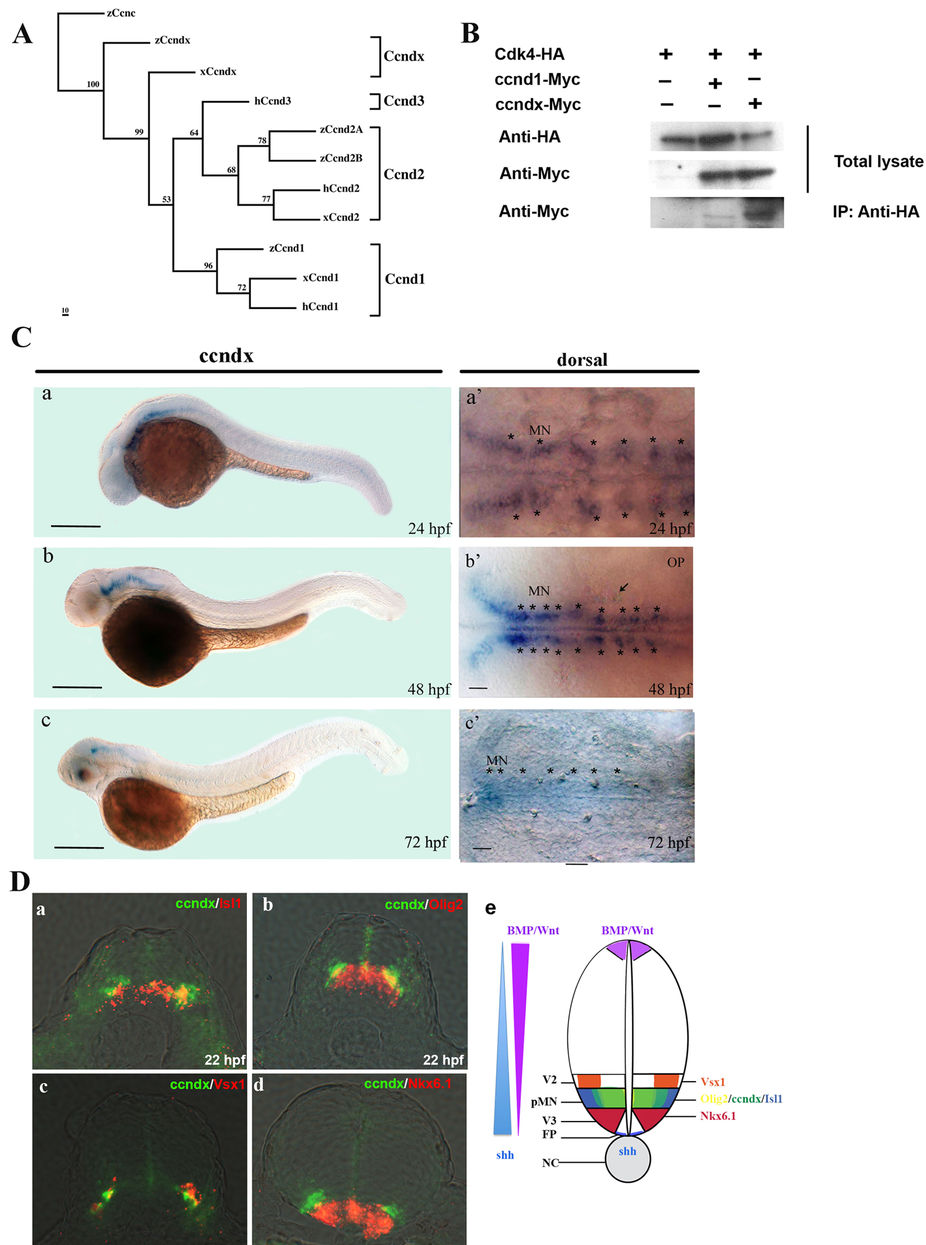
(A) Phylogenetic tree analysis of zccndx with other cyclin D proteins. Four groups of the cyclin D family, ccnD1, D2, D3, and Dx, exhibited perfect separation. The zccndx protein was grouped with Xenous ccndx. Analysis was performed using the parsimony method with 1000 bootstraps. (B) The zccndx protein binds to Cdk4. COS-1 cells were cotransfected with the indicated constructs, and cell lysates were co-immunoprecipitated (IP) with anti-HA monoclonal antibody; the anti-Myc monoclonal Ab was used for detection. The immunoblot was probed using the indicated antibodies. (C) At 24 hpf, zccndx mRNA was observed to be expressed in the motor neurons of hindbrain and spinal cord (a and a′). At 48 hpf, zccndx mRNA was also expressed in the hindbrain, but not in the spinal cord (b and b′). At 72 hpf, zccndx mRNA was weakly expressed in the hindbrain, with no signal in the spinal cord (c and c′). Dorsal views of zccndx mRNA signal in the hindbrain are shown (a′–c′). Scale bar, 100 mm. (D) Sections were made in the hindbrain at 22 hpf. The zccndx signal is shown in green and the isl1 (panel a), olig2 (panel b), and vsx1 (panel c), and nkx6.1 (panel d) mRNA signals are shown in red. The cartoon (panel e) shows the location of zccndx (green), isl1 (purple), olig2 (orange), vsx1 (blue), and nkx6.1 (red) signals. The Shh (blue) signaling gradient has been created from the floor plate to diffuse along the ventral tube while BMP/Wnt (purple) gradient shows opposite direction. The interaction of the dorsalizing and ventralizing signals defines the proper position of motoneurons. Scale bar, 100 mm.