Fig. S2
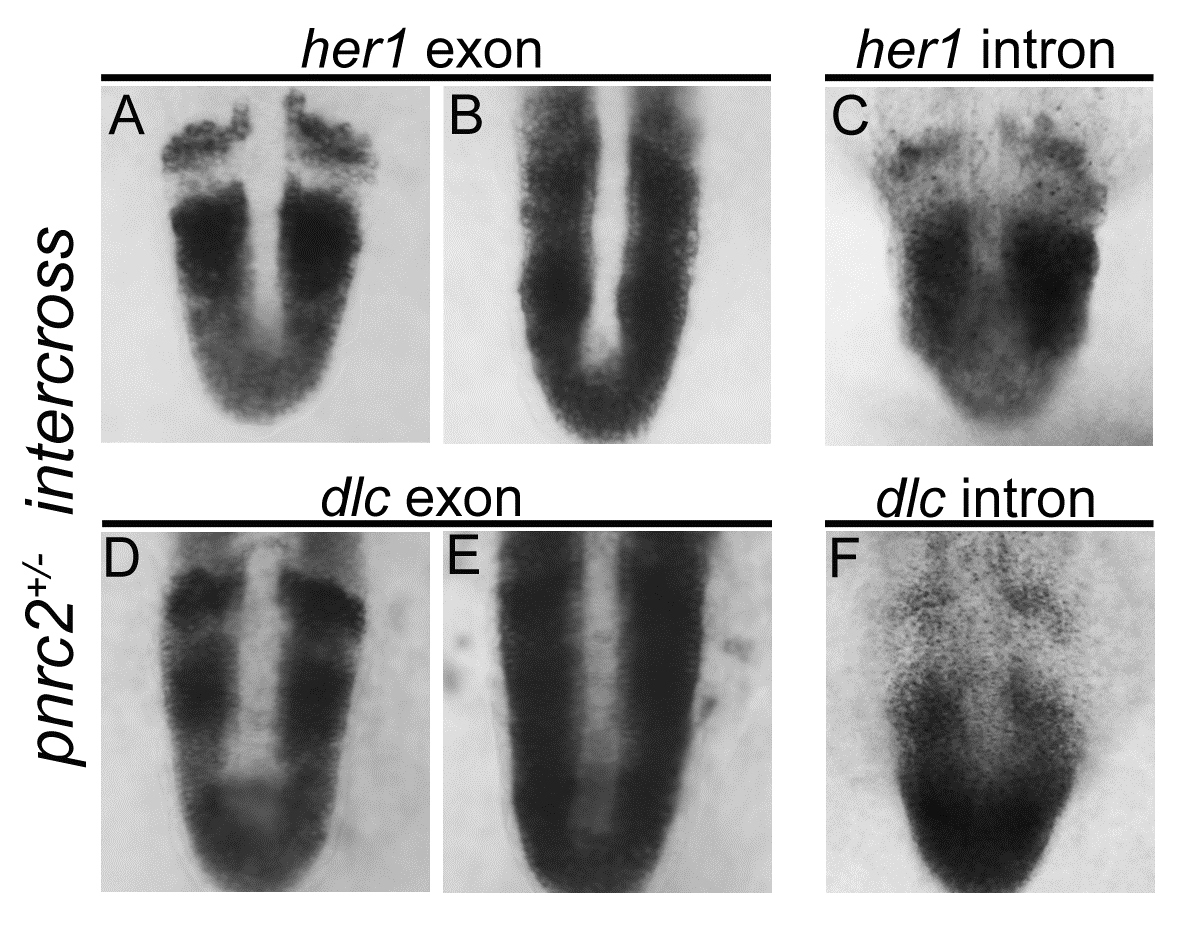
her1 and dlc transcripts accumulate post-transcriptionally in pnrc2oz22mutants. Exonic in situ probes reveal that segmentation clock-associated her1 and dlc transcripts are misexpressed in the expected one-quarter of embryos in a pnrc2oz22 intercross, n=13/57 (X2=0.1, p=0.7) and n=8/30 (X2=0.04, p=0.83), respectively (A, B and D, E). Intronic in situ probes, however, reveal no differences in expression among embryos from the same clutch, n=30/30 (X2=10.0, p=0.0016) and n=45/45 (X2=15.0, p=0.0001), respectively (C, F). These results are consistent with previous observations in torb644 mutants using intronic and exonic in situ probes that distinguish nascent from processed transcripts (Dill et al, 2005).
Reprinted from Developmental Biology, 429(1), Gallagher, T.L., Tietz, K.T., Morrow, Z.T., McCammon, J.M., Goldrich, M.L., Derr, N.L., Amacher, S.L., Pnrc2 regulates 3'UTR-mediated decay of segmentation clock-associated transcripts during zebrafish segmentation, 225-239, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.