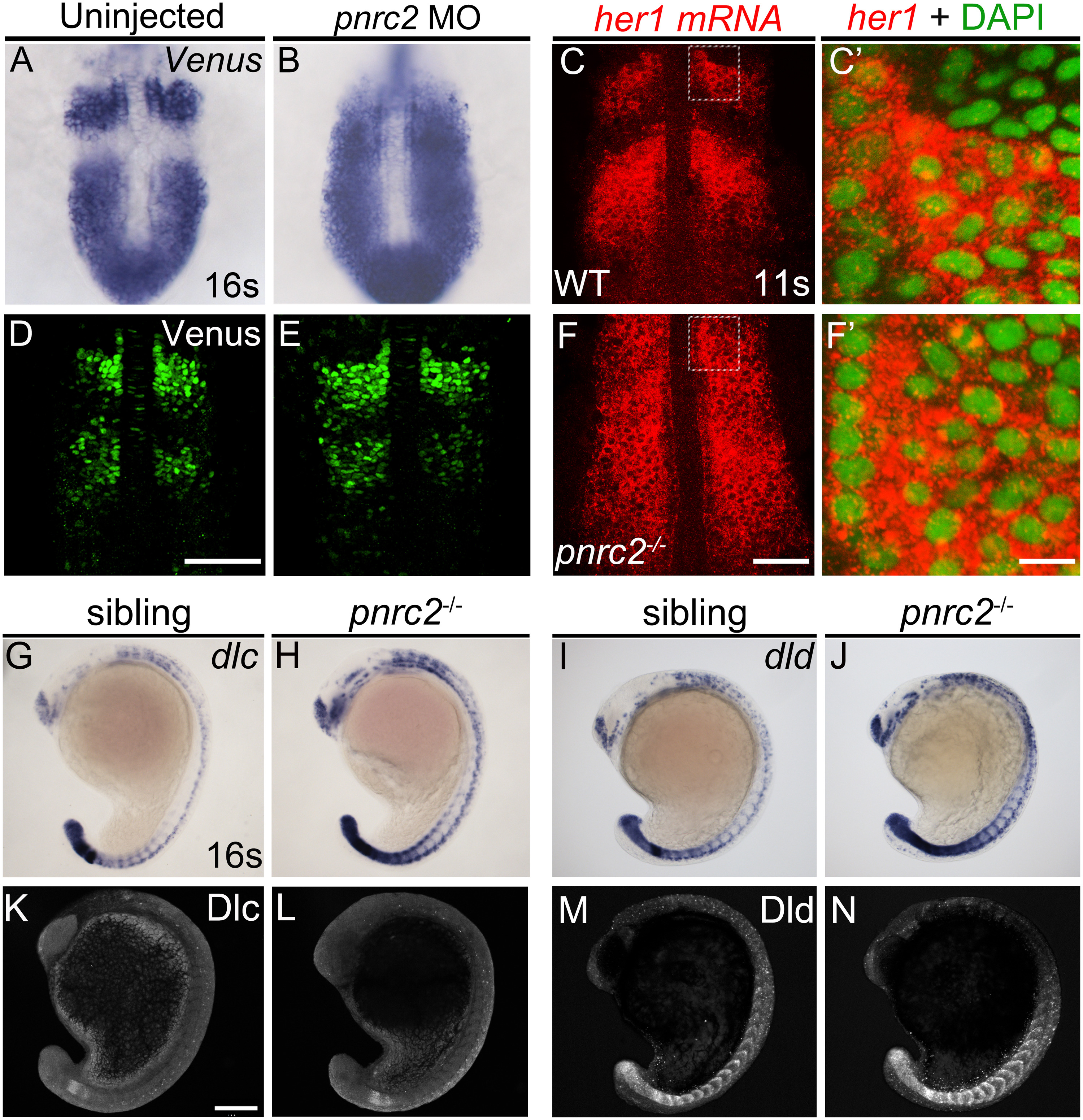
Fig. 7
Fig. 7 Both reporter and endogenous cyclic transcripts accumulate in Pnrc2-depleted embryos, but protein expression appears normal. Embryos carrying the her1:her1-Venusbk15transgenic clock reporter were injected with pnrc2 splice-blocking morpholino (sbMO) and processed to detect Venus transcripts (A, B) and Venus protein (D, E) at mid-segmentation stages. Representative embryos are shown in A (n=21/21); B (n=18/18), D (n=32/32), and E (n=29/29). Venus immunofluorescence panels (D, E) are at slightly higher magnification than Venus in situ panels (A, B). Detection of her1 mRNA by in situ hybridization chain reaction (HCR-ISH) (C, F) is consistent with chromogenic NBT/BCIP-based in situ detection of endogenous her1 transcript in wild-type and pnrc2 mutant embryos (Fig. 2F, H?), with substantial cytoplasmic localization revealed by DAPI counter staining in 500X magnified view (C?, F?). Because relative intensity of her1 HCR-ISH in pnrc2 mutants to wild-type embryos is high, levels have been reduced in pnrc2 mutant panels (F-F?; see Fig S6 A???, B???). Misexpression of dlc and dld mRNA is detected throughout the presomitic mesoderm (PSM), formed somites and neurons in the expected one-quarter of embryos in a pnrc2oz22intercross, n=5/28 (?2=0.76, p=0.4) and n=7/40 (?2=1.2, p=0.3), respectively (G-J). In contrast, Dlc and Dld protein expression is indistinguishable among siblings of the same pnrc2oz22 heterozygote intercross (K-N). Dlc and Dld immunolabeled embryos were genotyped prior to imaging and a subset of wild-type and mutant siblings were imaged by confocal microscopy with representative embryos shown (K-N). Total genotyped individuals per representative panel: n=5 (K), n=6 (L), n=12 (M), n=4 (N). Scale bars=50 Ám (D, F), 50 nm (F?), 100 Ám (K).
Reprinted from Developmental Biology, 429(1), Gallagher, T.L., Tietz, K.T., Morrow, Z.T., McCammon, J.M., Goldrich, M.L., Derr, N.L., Amacher, S.L., Pnrc2 regulates 3'UTR-mediated decay of segmentation clock-associated transcripts during zebrafish segmentation, 225-239, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.