Fig. 2
Fig. 2
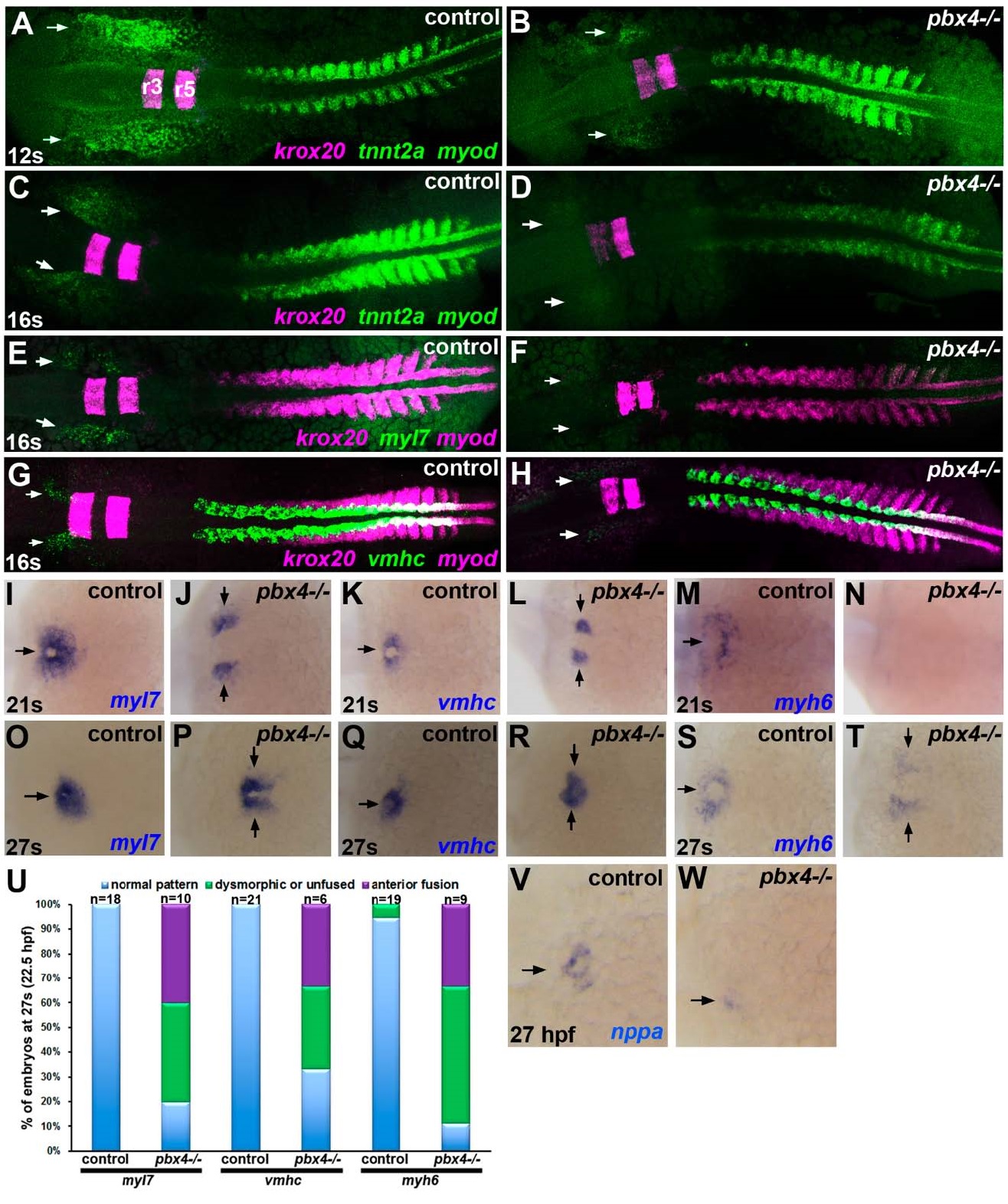
Delayed onset of cardiomyocyte differentiation genes in pbx4 mutant embryos. (A?T, V?W) RNA in situ expression of cardiomyocyte differentiation genes (A?D) tnnt2a, (E?F, I?J, O?P) myl7, (G?H, K?L, Q?R) vmhc, (M?N, S?T) myh6, and (V?W) nppa in (A,C,E,G,I,K,M,O,Q,S,V) control and (B,D,F,H,J,L,N,P,R,T,W) pbx4b557-/- embryos. Developmental stages are indicated. Embryos are shown in dorsal view, anterior towards the left. (A?H) Expression of the Pbx-dependent gene krox20, marking rhombomeres 3 and 5 (r3, r5 in A) in the hindbrain, was included to distinguish between control and pbx4b557-/- mutants [12,19]. Expression of myod, which is expressed in a stripe in each somite plus two additional pre-somitic stripes [38], was included for somite staging. Arrows indicate ALPM expression domains of cardiomyocyte differentiation genes. (A,B) At 12 s, tnnt2a is absent or reduced in pbx4b557-/- embryos (n = 8; 6/8 absent, 2/8 reduced) compared to controls (n = 30; all similar). (C,D) at 16 s, tnnt2a is reduced in pbx4b557-/- embryos (n = 7) compared to controls (n = 28). (E,F) at 16 s, myl7 is absent or reduced in pbx4b557-/- embryos (n = 9; 3/9 absent, 6/9 reduced) compared to controls (n = 22). (G,H) at 16 s, vmhc is absent or reduced in pbx4b557-/- embryos (n = 6; 1/6 absent, 5/6 reduced) compared to controls (n = 10). (I?T) At 21 s and 27 s, myl7, vmhc, and myh6 show delayed fusion and abnormal patterning of expression domains in pbx4b557-/- embryos compared to controls. For (I?N), numbers of affected embryos are provided in the text. For (O?T), numbers of affected embryos are graphed in (U). Arrows indicate expression domains of cardiomyocyte differentiation genes. (U) Graph displaying percentage of embryos at 27 s with either normally patterned (blue), dysmorphic or unfused (green), or anteriorly fused (purple) cardiac primordia. (V?W) At 27 hpf, nppa is reduced in pbx4b557-/- embryos (n = 4, all similar reduced expression) compared to controls (n = 24). Arrows indicate myocardial expression domains.