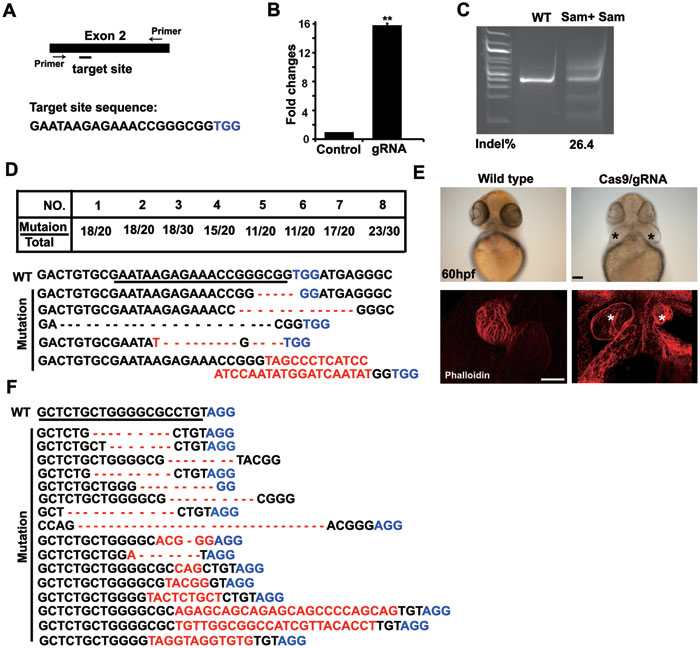
Fig. 3
Cas9/gRNA induces indels in the gata5 and gata4 loci in zebrafish. (A) Cartoon showing the position of the gRNA-targeting site and its sequence in the gata5 locus in zebrafish. (B) SSA recombination assay showing luciferase activity with and without cleavage of the target site by the Cas9/gRNA system in 293T cells. The luciferase activity increased by 16-fold in the group with gRNA (Cas9 + gRNA) compared with the control group (Cas9 + empty vector). Error bars indicate SD, n = 3. **P < 0.01. (C) Representative SURVEYOR assay showing the efficiency of Cas9-mediated cleavage, 26.4% in a single embryo at 50 hpf. (D) Representative Sanger sequencing results of the PCR amplicons of 8 individual embryos at 50 hpf, showing indels induced by Cas9/gRNA in the targeted gata5 locus. Twenty to thirty clones were sequenced for each embryo. Table summarizes the frequencies of site-specific indels, ranging from 50% (11/20) to 90% (18/20), in 8 individual embryos tested by Sanger sequencing. TGG (blue) is the PAM sequence. Deletion is represented by a dashed line and insertion is highlighted in red. (E) Two small hearts were formed in a Cas9/gRNA-induced mutant (right), which phenocopied that in the fautm236a genetic mutant, whereas a single heart formed in control embryos at 60 hpf. The heart was stained with phalloidin. * indicates two small hearts. Scale bar, 100 μm. (F) Representative Sanger sequencing results of the PCR amplicons of 8 individual embryos at 50 hpf, showing indels induced by Cas9/gRNA in the targeted gata4 locus as in D. AGG (blue) is the PAM sequence. AGG in 2 out of 16 sequencing results was deleted.