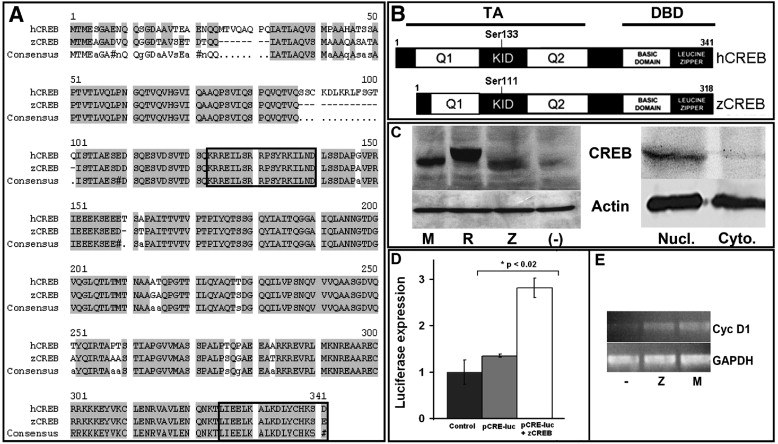
Fig. 1 Characterization of zebrafish CREB (zCREB) structure and function shows significant homology to human CREB. (hCREB; A) Alignment of hCREB (Gen Bank Accession No. NM_004379) and zCREB (BC044436) amino acid sequences showing 88.7% sequence identity. Boxes highlight the conserved epitopes recognized by antibodies targeted against pCREB (“KRRE” domain; residues 123–139) or total CREB (“LKDL” domain; residues 326–341). (B) Predicted protein domain structure and alignment shows zCREB is a protein of 311 amino acids, with conserved key regulatory domains (TA, Trans-Activation Domain; KID, Kinase Inducible Domain; Q1/Q2, Glutamine Rich Regions; DBD, DNA-Binding Domain). (C) Immunoblotting of protein lysates extracted from HEK-293 cells transiently-transfected with cDNAs encoding full-length mouse CREB (mCREB; M) FLAG-tagged rat CREB (R), zCREB (Z) or vehicle negative control (-) with anti-CREB “LKDL” antibody showing detection of all three CREB protein species (actin shown as loading control). Additionally, immunoblotting of nuclear (nucl.) or cytoplasmic (cyto.) protein extracts from 24 hpf zebrafish embryos shows that zCREB is a nuclear protein. (D) Luciferase reporter assay using a CREB-responsive luciferase reporter plasmid (pCRE-Luc) showing increased luciferase activity following transfection of HEK-293 cells with zCREB. (E) Semi-quantitative RT-PCR following transfection of HEK-293 cells with mCREB (M), zCREB (Z) or vehicle negative control (-) showing increased mRNA abundance of the direct CREB target gene cyclin D1, relative to housekeeping gene GAPDH.
Reprinted from Developmental Biology, 307(1), Dworkin, S., Heath, J.K., Dejong-Curtain, T.A., Hogan, B.M., Lieschke, G.J., Malaterre, J., Ramsay, R.G., and Mantamadiotis, T., CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish, 127-141, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.