- Title
-
SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos
- Authors
- Meshalkina, D.A., Glushchenko, A.S., Kysil, E.V., Mizgirev, I.V., Frolov, A.
- Source
- Full text @ Genes (Basel)
|
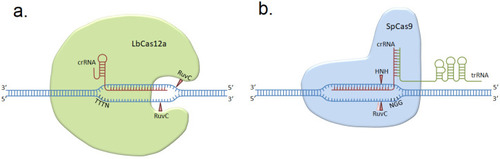
The mechanisms of action of LbCas12a ( |
|
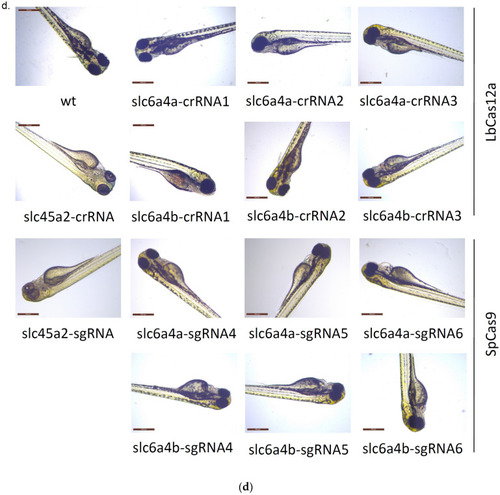
Assessment of crRNAs activities: ( |
|
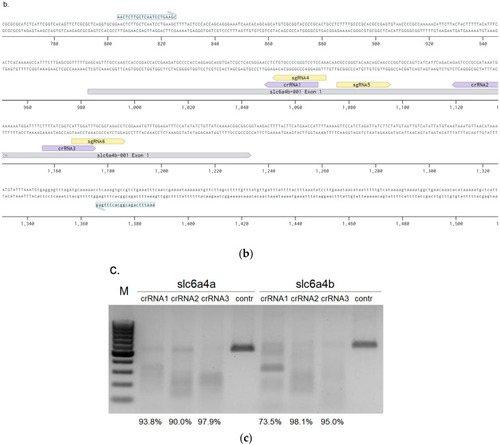
Assessment of crRNAs activities: ( |
|
Assessment of crRNAs activities: ( |
|
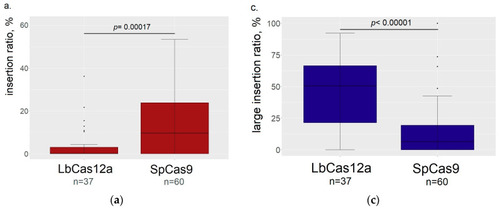
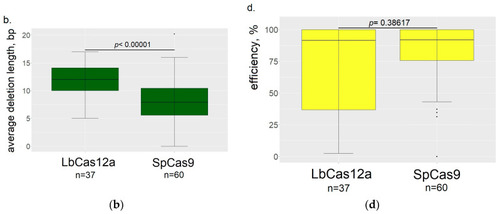
Comparison of LbCas12a and SpCas9 editing properties (the data from both serotonin transporters are summed together). ( |
|
Comparison of LbCas12a and SpCas9 editing properties (the data from both serotonin transporters are summed together). ( |