Fig. 2-S1
- ID
- ZDB-FIG-180417-21
- Publication
- Rogers et al., 2017 - Nodal patterning without Lefty inhibitory feedback is functional but fragile
- Other Figures
- All Figure Page
- Back to All Figure Page
|
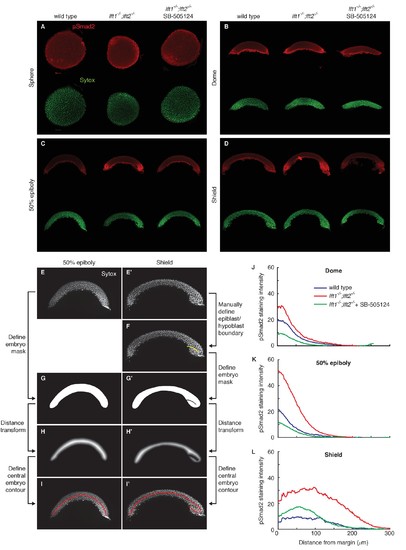
pSmad2 gradient quantification. pSmad2 gradients were visualized by α-pSmad2 immunofluorescence in wild type embryos, lft1-/-;lft2-/- mutant embryos, and lft1-/-;lft2-/- mutant embryos treated with 2 μM SB-505124 (Nodal inhibitor drug) starting at the 8 cell stage. (A–D) Representative maximum intensity projections for α-pSmad2 staining (red) and Sytox nuclear stain (green) in sphere (A), dome (B), 50% epiboly (C), and shield (D) stage embryos. Sphere stage embryos are animal pole views; all other stages are laterally oriented. For lateral images, projections were derived from the 10 slices surrounding the center of the embryo. (E–I’) Nodal signaling gradients were quantified by measuring nuclear pSmad2 immunofluorescence as a function of distance from the margin. To quantify gradients, individual nuclei were first identified, their distance from the embryo margin along a contour following the midline of the embryo cross-section was measured, the fluorescence intensity in each nucleus was determined, and finally intensity versus distance was plotted (see Materials and methods). The process of defining the contour for 50% epiboly (E,G,H,I) and shield (E’,F’,G’,H’,I’) stage embryos is illustrated here. (E,E’) Maximum intensity projections of Sytox green stacks used to identify nuclei and define embryo contour. The ten planes closest to the center of the embryo (five planes above and five below center) were used for quantification and calculation of the central contour. (F) For shield stage embryos, the boundary between the epiblast and involuting hypoblast was manually defined (yellow line). This boundary allowed the central contour to ‘wrap’ around the epiblast/hypoblast boundary at the embryonic shield. (G,G’) A minimal mask containing all segmented nuclei was defined. For shield stage embryos, pixels corresponding to the hypoblast/epiblast boundary were set to zero. (H,H’) To identify an approximate embryo midline, the distance transform of the inverse of the embryo mask (panels G,G’) was calculated. In this new representation, pixels within the mask that are the farthest from the mask boundary (i.e. pixels along its midline) have the highest values. (I,I’) The central contour of each embryo was defined by stepping from one margin to the other along the central maxima of the distance transform. The final contours used for gradient quantification are shown in red. (J–L) pSmad2 gradients are plotted for dome (J), 50% epiboly (K), and shield stages (L). Gradients in lft1-/-;lft2-/- mutants (red) have higher amplitudes than wild type (blue) or drug-treated lft1-/-;lft2-/- mutant (green) gradients. Some images from (A), (C), and (D) are also used in Figure 2. |
| Antibody: | |
|---|---|
| Fish: | |
| Condition: | |
| Anatomical Term: | |
| Stage Range: | Sphere to Shield |
| Fish: | |
|---|---|
| Observed In: | |
| Stage Range: | Sphere to 50%-epiboly |